Based on publications, and responses from reviewers on the subject, it is evident that neutral evolution is a misused and misunderstood concept in evolutionary computation circles. I intend to dispel some of the confusion in this series of post based on some material from a journal paper by Dr. Kaur (my doctoral supervisor) and myself [i].
Background
Kimura’s neutral theory of evolution [ii] hypothesizes that most genetic changes are neutral (i.e. they do not affect fitness); it also states that most evolutionary changes are the result of random genetic drift among networks of neutral genomes.
Evolutionary Computing practitioners have tried to simulate this situation by using degenerate genotype-to-phenotype maps. A map is degenerate when more than one genotype expresses the same phenotype. Neutral Evolution is evolution using such a degenerate map such that there are not only changes that improve the individuals, but there is also the possibility of random “neutral” drifts that don’t affect the fitness of individuals.
The subset of Evolutionary Computing literature related to genetic algorithms inconsistently use redundancy as a synonym for degeneracy (e.g. [iii], [iv], [v]). Within genetic programming we see the use of such similar concepts as functional redundancies [vi], [vii], and redundancies due to gene not used in mapping the genome to its phenotype. The paper goes into more details on the sometimes confusing use of terms related to neutral evolution.
Review
Because Kimura’s work and other biology-based evolutionary studies (e.g. [viii], [ix]) suggested that neutrality plays some role in evolutionary search, the Evolutionary Computing community has been interested in analyzing and utilizing its effects. The majority of works have been devoted to empirically exploring the relation between using some degenerate code and performance.
Degenerate code have been reported to give better (or at least equal) performance to nondegenerate codes in many works; these include Harvey and Thompson [x], Julstrom [xi], Banzhaf [xii], Keller and Banzhaf [xiii], Yu and Miller [xiv],[xv], Shackleton et al. [xvi], Shipman et al. [xvii] and Ebner et al. [xviii]. In such cases of performance improvement it is usually argued that using a degenerate code helped in avoiding premature convergence and allows escape from local optima on saddle surfaces.
Some works have found using a degenerate code caused a deterioration of performance; these include Davis [xix], Eschelman and Schaffer [xx], Ronald et al. [xxi], Smith et al. [xxii],[xxiii], and Knowles and Watson [xxiv]. Reasons advanced for low performance include loss of genetic diversity, different genotypes that represent the same phenotype competing against each other and a larger search space size.
A common methodology used to determine whether or how using some degenerate code affects search is a fairly frequent approach in computational intelligence circles; the approach is to apply your algorithm (both with and without using degeneracy) to a set of “representative” problems and compare the resulting performance. This methodology is at best weak, especially when it is not supported by any serious analytical work. For one thing the way various factors interact in an evolving populations are usually too complex to infer how that population evolved based on simply comparing performance (which, however you measure it, is a summary statistic). This is partly responsible for the confused picture on the usefulness of neutral evolution.
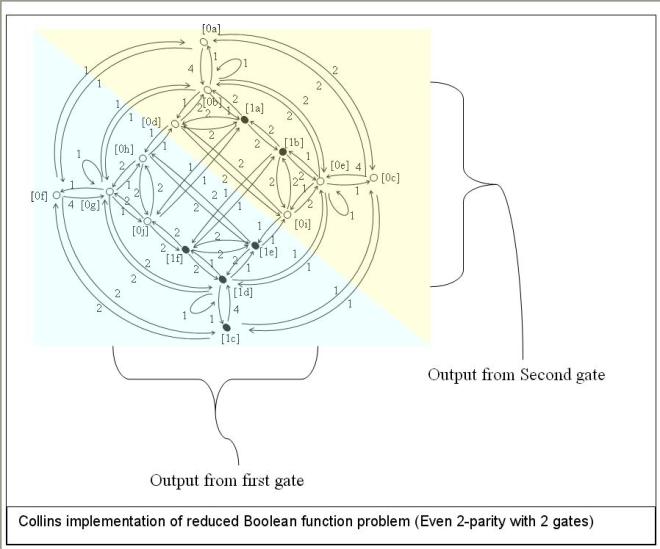
Galvan and Poli [xxv], [xxvi] did quite a nice job of detailing reasons why results on neutrality are mixed. Apart from the inconsistent definitions and methodological issues already alluded to, they also pointed out that the features of a problem can change with adding neutrality. Examples of such changes are graphically detailed using quotient graphs in our paper.
Conclusion
In our (i.e. Dr Kaur and me) paper on “Search, Neutral Evolution and Mapping”, we showed specific cases when using a degenerate code does not affect search performance. For other cases there have been published reports of both successes (i.e. performance improvement) and failures on using degenerate codes. The published successes outnumber the unsuccessful outcomes. My guess is that the imbalance is due to two biases:
- Researchers are more likely to publish their successful results;
- Reviewers are more likely to favourably review papers with successful outcomes;
I suspect however that the true number of performance improvements and deteriorations on using a degenerate code are quite close.
It is known that just playing with parameters (such as the mutation rate) can change whether using a degenerate code is more, or less successful. For that reason we resisted the temptation of reporting whether using degenerate coding was more or less successful in the experiments in the paper. Such a result is not general (and in our estimation not very useful).
Neutral Evolution is a coding issue and should be treated as such. Evolutionary computing practitioners should treat it in the same way other coding issues are treated and not expect to find a silver bullet in its use.
References
[i] D. Wilson, D Kaur, Search, Neutral Evolution and Mapping in Evolutionary Computing: A Case Study of Grammatical Evolution. IEEE Trans. Evol. Comp. pp 566-590, Vol. 13, issue 3, June 2009.
[ii] M. Kimura, Neutral theory of molecular evolution. Cambridge University Press, 1983.
[iii] J. Cohoon, S. Hegde, W. Martin and D. Richards, “Floorplan design using distributed genetic algorithms”, Computer-Aided Design, ICCAD-88. Digest of Technical Papers., IEEE International Conference, pp. 452-455, 1988.
[iv] S Ronald, J Asenstorfer and M Vincent “Representational Redundancy in Evolutionary Algorithms” , Proceedings of the 1995 IEEE International Conference on Evolutionary Computing, Volume 2, pg 631-637. 1995.
[v] N. Radcliffe and P. Surry, “Fitness Variance of Formae and Performance Prediction”. FOGA3, pp.51-72. 1994.
[vi] J Miller and P. Thomson, “Cartesian Genetic Programming” Proceedings of the 3rdEuropean Conferenceon Genetic Programming. LNCS Vol. 1802 pp. 121-132. 2000.
[vii] J. Miller and S. Smith, “Redundancy and Computational Efficiency in Cartesian Genetic Programming”, IEEE Transactions on Evolutionary Computation, Vol. 10, No. 2, pp. 167-174, 2006.
[viii] Fontana, W., Schuster, P. “Continuity in evolution: On the nature of transitions”. Science 280, pp. 1431–1433, 1998.
[ix] Huynen, M.A., “Exploring phenotype space through neutral evolution”. Molecular Evolution vol. 43, pp. 165–169, 1996.
[x] I. Harvey and A. Thompson. Through the labyrinth evolution finds a way: A silicon ridge. In Proceedings of the First International Conference on Evolvable Systems: From Biology to Hardware (ICES), pages 406–422. Springer-Verlag, 1996.
[xi] Julstrom, B. A., “Redundant genetic encodings may not be harmful”,Banzhaf, W. et al. , editors, GECCO99-1, Morgan Kaufmann Publishers, San Francisco, CA, pp.791, 1999.
[xii] W. Banzhaf. Genotype-phenotype-mapping and neutral variation – A case study in genetic programming. In Y. D. et al., editor, Parallel Problem Solving from Nature III, volume 866 of LNCS, pages 322–332, Jerusalem, 9-14 Oct. 1994. Springer-Verlag.
[xiii] R. E. Keller and W. Banzhaf. Genetic programming using genotype-phenotype mapping from linear genomes into linear phenotypes. In J. R. Koza, D. E. Goldberg, D. B. Fogel, and R. L. Riolo, editors, Genetic Programming 1996: Proceedings of the First Annual Conference, pages 116–122, Stanford University, CA, USA, 28–31 July 1996. MIT Press.
[xiv] T. Yu and J. Miller. Neutrality and the evolvability of boolean function landscape. In Fourth European Conference on Genetic Programming, pages 204–211. Springer-Verlag, 2001.
[xv] T. Yu and J. F. Miller. Needles in haystacks are not hard to find with neutrality. In J. A. Foster, E. Lutton, J. Miller, C. Ryan, and A. G. B. Tettamanzi, editors, Genetic Programming, Proceedings of the 5th European Conference, EuroGP 2002, volume 2278 of LNCS, pages 13–25, Kinsale, Ireland, 3-5 Apr. 2002. Springer-Verlag.
[xvi] M. Shackleton, R. Shipman, and M. Ebner. An investigation of redundant genotype-phenotype mappings and their role in evolutionary search. In Proceedings of the 2000 Congress on Evolutionary Computation CEC00 La Jolla Marriott Hotel pp. 493–500, 2000.
[xvii] R. Shipman, M. Shackleton, and I. Harvey. The use of neutral genotype-phenotype mappings for improved evolutionary search. BT. Technology Journal, 18(4):103–111, October. ISSSN 2000.
[xviii] M. Ebner, M. Shackleton and R. Shipman, “How Neutral Networks Influence Evolvability”. Complexity, 7(2), pp. 19-33, Wiley Periodicals, 2002.
[xix] L. Davis, Adapting operator probabilities in genetic algorithms. In Schaffer, J. D. (Ed.), Proceedings of the Third International Conference on Genetic Algorithms (pp. 61–69). San Mateo, CA: Morgan Kaufmann. 1989.
[xx] L. J. Eshelman, and J. D. Schaffer. Preventing premature convergence in genetic algorithms by preventing incest. In Belew, R. K., & Booker, L. B. (Eds.), Proceedings of the Fourth International Conference on Genetic Algorithms (pp. 115–122). San Mateo, CA: Morgan Kaufmann. 1991.
[xxi] S. Ronald, J. Asenstorfer, and M. Vincent, Representational redundancy in evolutionary algorithms. In 1995 IEEE International Conference on Evolutionary Computation, Vol. 2 pp. 631–636. 1995 .Piscataway, NJ.
[xxii] T. Smith, P. Husbands, and M. O’Shea. Neutral networks and evolvability with complex genotype-phenotype mapping. Lecture Notes in Computer Science, 2159:272–282, 2001.
[xxiii] T. Smith, P. Husbands, and M. O’Shea. Neutral networks in an evolutionary robotics search space. In Congress on Evolutionary Computation: CEC 2001, pages 136–145. IEEE Press, 2001.
[xxiv] J. D. Knowles and R. A. Watson, On the utility of redundant encodings in mutation-based evolutionary search. In Merelo, J. J., Adamidis, P., Beyer, H.-G., Fernandez-Villacanas, J.- L., & Schwefel, H.-P. (Eds.), Parallel Problem Solving from Nature, PPSN VII (pp. 88–98), Berlin: Springer-Verlag, 2002.
[xxv] E. Galvan-Lopez and R. Poli, “An Empirical Investigation of How and Why Neutrality Affects Evolutionary Search” GECCO’06, Seattle, Washington, USA.July 8–12, 2006,
[xxvi] E. Galván López and R. Poli. Some Steps Towards Understanding How Neutrality Affects Evolutionary Search, in T. P. Runarsson, H. Beyer, E. K. Burke, J. J. Merelo Guervos, L D. Whitley and X. Yao, editors, PPSN IX: Proceedings of the 9th International Conference on Parallel Problem Solving from Nature, volume 4193 of LNCS, pages 778 – 787, Reykjavik, Iceland, 9 – 13 September 2006. Springer.












![Reblog this post [with Zemanta]](https://i0.wp.com/img.zemanta.com/reblog_e.png)